The analysis, done in three colon cancer cell lines, is believed to be the first to profile transcriptome-level changes in response to DNA damage across individual cells.
11:00 AM
Author |
Chemotherapy works by attacking rapidly dividing cells within the body. But small pockets of cancer cells can withstand its assault, allowing the cancer eventually to return.
Gaining a better understanding of why some cancer cells survive while others die is critical for making chemotherapy more effective, says Jun Hee Lee, Ph.D., a cancer researcher at the University of Michigan Rogel Cancer Center.
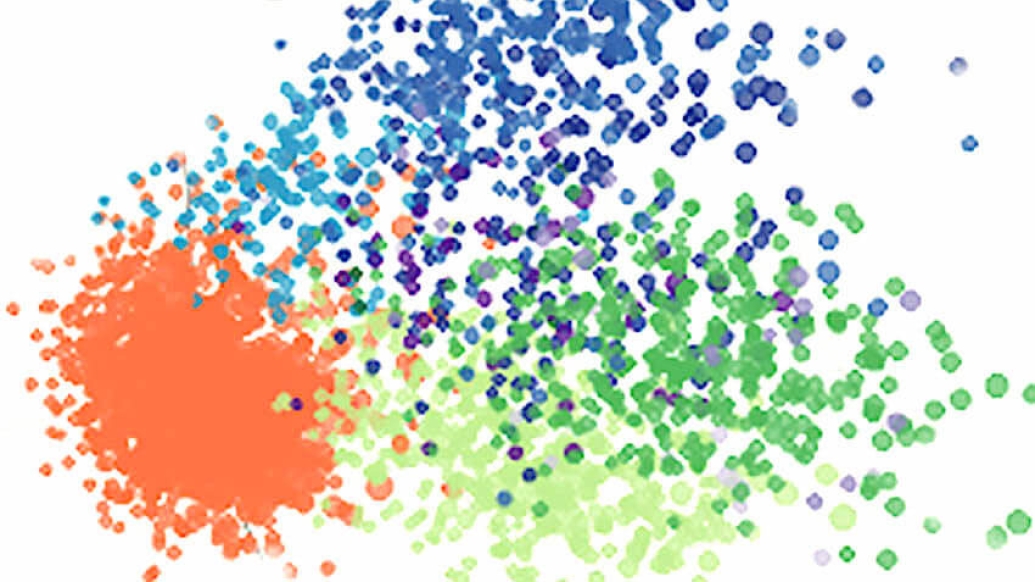
Using a technique called single-cell RNA sequencing, a research team from U-M was able to show for the first time how individual cells within a single population of cancer cells respond differently to the DNA damage caused by chemotherapy. The responses, they found, fall into three groups: activating genes that control cell death, cell division, or stress response, according to findings published in Cell Reports.
LISTEN UP: Add the new Michigan Medicine News Break to your Alexa-enabled device, or subscribe to our daily updates on iTunes, Google Play and Stitcher.
"Collectively, we observed that cells with different fates actually had completely distinct sets of activated genes and that these different 'transcriptomic landscapes' dictate the fates of cells after DNA damage from chemotherapy," says Lee, co-senior author of the study and an associate professor of molecular and integrative physiology at Michigan Medicine.
While DNA contains the complete instruction manual for the cell, sequences that are transcribed into RNA tell the story of which genes are switched on or off at a given time — that is, which sets of individual instructions are being acted upon. The transcriptome is the complete set of these RNA sequences within a given cell.
Applying single-cell techniques
Among scientists, single-cell analysis is frequently compared to a fruit smoothie, Lee notes. Many types of studies measure characteristics or responses across a group of cells — a mixture of individual players that contribute to a greater whole, like fruit in a smoothie. This can provide useful information, but can also obscure differences between and among the individual contributors — the cell-level equivalent of the strawberries and blueberries and bananas in the smoothie. Single-cell techniques allow those individual differences to be teased out.
If we learn that there's a certain sub-population of cells with specific characteristics that allow them to survive chemotherapy when other cells die, then scientists might look for ways to target those cells specifically.Jun Hee Lee, Ph.D.
The study analyzed more than 10,000 cells from three colon cancer cell lines. The cells were exposed to the chemotherapy agent fluorouracil, which is commonly used against colon cancer and other types of cancer. Some of the observations were replicated with additional techniques and different chemotherapy drugs.
"Previously, the scientific consensus was that DNA damage leads to a fairly uniform transcriptional response, which leads to different cell fates in a passive way, based on the given levels of gene expression in the cell," Lee says. "In contrast, we found that different DNA damage response genes were often upregulated only in the subset of cells that all share a particular cell fate."
The group is conducting ongoing research to understand what factors that cause some cells have one fate and others to have a different fate.
MORE FROM MICHIGAN: Sign up for our weekly newsletter
"If we learn that there's a certain sub-population of cells with specific characteristics that allow them to survive chemotherapy when other cells die, then scientists might look for ways to target those cells specifically," Lee says
Making data available to other researchers
The research team, which was co-led by Hyun Min Kang, Ph.D., an associate professor of biostatistics at the School of Public Health, is also making their data available online for other researchers.
"For instance, other scientists can examine how individual genes are expressed across single cells before and after chemotherapy treatments. and how the specific gene expression is correlated with the chemotherapy dose, or with the expression of other genes," Kang says. "The online tool can also be used by researchers to test new hypotheses and generate new data — and therefore has the potential to accelerate future research on DNA damage responses."
This work was supported by grants from the National Institutes of Health (R01DK102850, R01DK114131, U01HL137182, P30AG024824, P30DK034933, P30DK089503, P30CA046592), the Chan Zuckerberg Initiative, U-M's MCubed Initiative, an Organogenesis
Fellowship and an American Association for the Study of Liver Diseases Pilot Research Award.
Additional authors on the study include: Sung Rye Park, Leon Friesen, Chun-Seok Cho, Zac Zezhi Zhang, Yu-Chih Chen and Chang H. Kim of U-M; Sim Namkoong of U-M and Kangwon National University, Korea; Euisik Yoon of U-M and Yonsei University, Korea; and Hojoong Kwak of Cornell University.
Paper cited: "Single cell transcriptome analysis of colon cancer cell response to 5-fluorouracil-induced DNA damage," Cell Reports. DOI: 10.1016/j.celrep.2020.108077

Explore a variety of healthcare news & stories by visiting the Health Lab home page for more articles.

Department of Communication at Michigan Medicine
Want top health & research news weekly? Sign up for Health Lab’s newsletters today!